Module 2: Data Preparation using the Tidyverse
Packages in R
Role of Packages in R
Packages in R are similar to user-written commands (think ssc install) in Stata.
But most things you do in Stata probably use core Stata commands.
In R, most of your analysis will probably be done using packages.
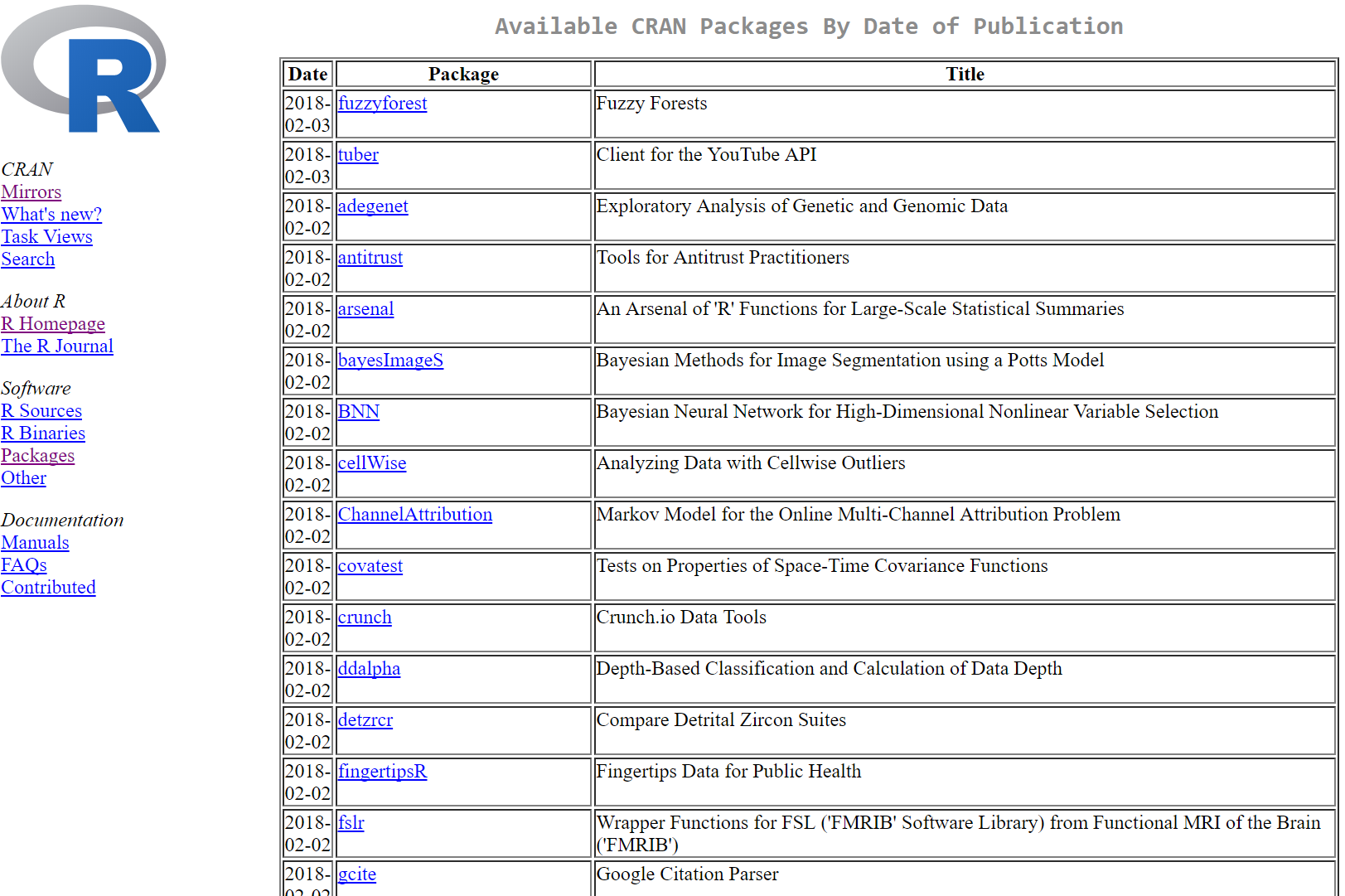
Installing and using a package
To install a package, use the function (preferably in the console) install.packages()
- To begin with, let’s install 2 packages:
install.packages("tidyverse") # Install tidyverse
install.packages("rio") # Install rioLoading a package during analysis
Unlike Stata, in R you need to declare what packages you will be using at the beginning of each R document.
To do this, use the library() function.
-require() also works, but its use is discouraged for this purpose.
library("tidyverse") # Install tidyverse
library("rio") # Install rio
Data Prep Preliminaries
Import and export using rio
Previously, importing and exporting data was a mess, with a lot of different functions for different file formats:
- Stata DTA files alone required two functions: read.dta (for Stata 6-12 DTA files), read.dta13 (for Stata 13 and later files), etc.
The rio package simplifies this by reducing all of this to just one function, import()
- Automatically determines the file format of the file and uses the appropriate function from other packages to load in a file.
PISA_2015 <- import("data/PISA2015.sas7bdat")
PISA_2015[1:5,1:6]## CNTRYID CNT CNTSCHID CYC NatCen Region
## 1 8 ALB 800001 06MS 000800 800
## 2 8 ALB 800002 06MS 000800 800
## 3 8 ALB 800003 06MS 000800 800
## 4 8 ALB 800004 06MS 000800 800
## 5 8 ALB 800005 06MS 000800 800export(PISA_2015, "PISA_2015.rds")Tibbles: an update to the data frame
Last class, we covered data frames—the most basic data object class for data sets with a mix of data class.
Today, we introduce one final data object: the tibble!
The tibble can be thought of as an update to the data frame—and it’s the first part of the tidyverse package that we’ll look at.
Tibble vs data frames
There are three main benefits to the tibble:
- Displaying data frames:
- If you display a data frame, it will print as much as much output as allowed by the “max.print” option in the R environment. With large data sets, that’s far too much. Tibbles by default print the first 10 rows and as many columns as will fit in the window.
- Partial matching in data frames:
- When using the $ method to reference columns of a data frame, partial names will be matched if the reference isn’t exact. This might sound good, but the only real reason for there to be a partial match is a typo, in which case the match might be wrong.
- Tibbles are required for some functions.
Creating or converting to tibbles
The syntax for creating tibbles exactly parallels the syntax for data frames:
tibble() creates a tibble from underlying data or vectors.
as_tibble() coerces an existing data object into a tibble.
PISA_2015 <- as_tibble(PISA_2015); PISA_2015[1:5,1:5]## # A tibble: 5 x 5
## CNTRYID CNT CNTSCHID CYC NatCen
## <dbl> <chr> <dbl> <chr> <chr>
## 1 8 ALB 800001 06MS 000800
## 2 8 ALB 800002 06MS 000800
## 3 8 ALB 800003 06MS 000800
## 4 8 ALB 800004 06MS 000800
## 5 8 ALB 800005 06MS 000800Glimpse
Another tidyverse function that’s very useful is glimpse() , a function very similar to str().
Both functions display information about the structure of a data object.
str() provides more information, such as column (variable) attributes embedded from external data formats, but consequently is much less readable for complex data objects.
glimpse() provides only column names, classes, and some data values (much more readable)
I will often use str() when I want more detailed information about data structure, but use glimpse() for quicker glances at the data.
Pipes
Another major convenience enhancement from the tidyverse is pipes, denoted %>%,
Pipes allow you to combine multiple steps into a single piece of code.
Specifically, after performing a function in one step, a pipe takes the data generated from the first step and uses it as the data input to a second step.
Pipes Example
barro.lee.data <- import("data/BL2013_MF1599_v2.1.dta") %>%
as_tibble() %>% glimpse(width = 50)## Observations: 1,898
## Variables: 20
## $ BLcode <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, …
## $ country <chr> "Algeria", "Algeria", "Alge…
## $ year <dbl> 1950, 1955, 1960, 1965, 197…
## $ sex <chr> "MF", "MF", "MF", "MF", "MF…
## $ agefrom <dbl> 15, 15, 15, 15, 15, 15, 15,…
## $ ageto <dbl> 999, 999, 999, 999, 999, 99…
## $ lu <dbl> 80.68459, 81.05096, 82.6111…
## $ lp <dbl> 17.563400, 17.018442, 14.31…
## $ lpc <dbl> 3.745905, 3.464397, 3.06939…
## $ ls <dbl> 1.454129, 1.639253, 2.75251…
## $ lsc <dbl> 0.4595877, 0.4952279, 1.049…
## $ lh <dbl> 0.2978759, 0.2594140, 0.322…
## $ lhc <dbl> 0.16479027, 0.14177565, 0.1…
## $ yr_sch <dbl> 0.8464569, 0.8350149, 0.880…
## $ yr_sch_pri <dbl> 0.7443995, 0.7284052, 0.706…
## $ yr_sch_sec <dbl> 0.09280412, 0.09858591, 0.1…
## $ yr_sch_ter <dbl> 0.009253278, 0.008023791, 0…
## $ pop <dbl> 5241, 5699, 6073, 6374, 710…
## $ WBcode <chr> "DZA", "DZA", "DZA", "DZA",…
## $ region_code <chr> "Middle East and North Afri…
Data Preparation
Tidyverse and the verbs of data manipulation
A motivating principle behind the creation of the tidyverse was the language of programming should really behave like a language.
Data manipulation in the tidyverse is oriented around a few key “verbs” that perform common types of data manipulation.
- filter() subsets the rows of a data frame based on their values.
- select() keeps variables (columns) based on their names.
- mutate() adds new variables that are functions of existing variables.
- summarize() creates a number of summary statistics out of many values.
- arrange() changes the ordering of the rows.
Note: the first argument for each these functions is the data object (so pipe!).
Filtering data
Filtering keeps observations (rows) based on conditions.
- Just like using use subset conditions in the row arguments of a bracketed subset
# Using brackets
wages[(wages$schooling > 10) & (wages$exper > 10),] ## wage schooling sex exper
## 2 249.6774 13 female 11# Using filter
wages %>% filter(schooling > 10,exper > 10) ## wage schooling sex exper
## 1 249.6774 13 female 11Notice a couple of things about the output:
- It doesn’t look like we told filter() what data set we would be filtering.
- That’s because the data set has already been supplied by the pipe. We could have also written the filter as:
filter(wages, schooling > 10,exper > 10) ## wage schooling sex exper
## 1 249.6774 13 female 11- We didn’t need to use the logical &. Though multiple conditions can still be written in this way with filter(), the default is just to separate them with a comma.
Selecting data
Just like filter is in many ways a more convenient form of writing out bracketed row subset conditions, the verb select() is largely a more convenient method for writing column arguments.
# Using brackets
wages_row1[,c("wage","schooling","exper")]## wage schooling exper
## 1 134.2306 13 8# Using select
wages_row1 %>% select(wage,schooling,exper) ## wage schooling exper
## 1 134.2306 13 8An example of dropping a column
One option we have not covered so far in creating subsets is dropping rows or columns.
R has a specific notation for this, easily used with select():
wages_row1 # What wages_row1 looks like:## wage schooling sex exper
## 1 134.2306 13 female 8wages_row1 %>% select(-exper) #drop exper## wage schooling sex
## 1 134.2306 13 femaleDropping columns (or rows) using the - notation also works with brackets, but only when using the number location of the row or column to be dropped.
wages_row1[,-4] # works## wage schooling sex
## 1 134.2306 13 female# wages_row1[,-"exper"] does not workBecause of the ability to use name arguments, dropping with select() is generally easier.
“Mutating” data
Creating new variables that are functions of existing variables in a data set can be done with mutate().
mutate() takes as its first argument the data set to be used and the equation for the new variable:
wages <- wages %>%
mutate(expsq = exper^2) # Create expersq
wages # Display wages## wage schooling sex exper expsq
## 1 134.23058 13 female 8 64
## 2 249.67744 13 female 11 121
## 3 53.56478 10 female 11 121Summarizing data
Summary statistics can also be easily created using the tidyverse function summarize()
The summarize functions uses summary statistic functions in R to create a new summary tibble, with syntax largely identical to mutate().
Let’s try summarizing with the mean() summary statistic.
wages %>%
summarize(avg_wage = mean(wage))## avg_wage
## 1 145.8243Summary Statistics functions in R
There are a number of summary statistics available in R, which can be used either with the summarize() command or outside of it:
Measures of central tendency and spread:
Position:
Count:
Multiple summary variables
Let’s look at an example of using multiple summary variables with a larger 50-observation sample for the wages data set.
wages %>%
summarize(avg.wage = mean(wage), sd.wage = sd(wage),
avg.exper = mean(exper), sd.exper = sd(exper))## # A tibble: 1 x 4
## avg.wage sd.wage avg.exper sd.exper
## <dbl> <dbl> <dbl> <dbl>
## 1 5942. 17526. 7.47 2.08Grouping data
Creating summary statistics by group is another routine task. This is accommodated in the tidyverse using the group_by().
- The arguments of group_by(), in addition to the data set, are simply the grouping variables separated by commas.
wages %>% group_by(sex) %>%
summarize(avg.wage = mean(wage), sd.wage = sd(wage))## # A tibble: 2 x 3
## sex avg.wage sd.wage
## <fct> <dbl> <dbl>
## 1 female 5473. 18883.
## 2 male 6410. 16711.Arranging (sorting) data
If you want to sort your data by the values of a particular variable, you can easily do so as well with the arrange()
function.
wages[1:3,] %>% arrange(exper)## # A tibble: 3 x 4
## wage schooling sex exper
## <dbl> <int> <fct> <int>
## 1 175. 12 female 5
## 2 103. 11 male 7
## 3 1411. 14 female 8Not: arrange() sorts values in ascending order by default. If you want to sort in descending order, wrap the variable name inside desc() in the function.
Sampling from data
Creating a sample from a data set in R is made easy by two main function in R: sample_n and sample_frac.
Syntax:
sample_n(data, size, replace = FALSE/TRUE)
sample_frac(data, size = 1, replace = FALSE/TRUE)
A data prep example with fuel economy data
Let’s use tidyverse data manipulation verbs to work through a practical data prep problem from start to finish.
For the problem, Let’s use fuel economy data again, but with half of the data set. The data comes from the vehicles data set in the fueleconomy package.
# install.packages("fueleconomy") # Run only once
library(fueleconomy)Now let’s look at how fuel efficiency has changed over time in the data set. Specifically, let’s create descriptive statistics of fuel efficiency by year for “normal” passenger vehicles (4-8 cylinders).
What’s in the data set?
glimpse(vehicles[2:12], width=50)## Observations: 33,442
## Variables: 11
## $ make <chr> "AM General", "AM General", "AM G…
## $ model <chr> "DJ Po Vehicle 2WD", "DJ Po Vehic…
## $ year <int> 1984, 1984, 1984, 1984, 1985, 198…
## $ class <chr> "Special Purpose Vehicle 2WD", "S…
## $ trans <chr> "Automatic 3-spd", "Automatic 3-s…
## $ drive <chr> "2-Wheel Drive", "2-Wheel Drive",…
## $ cyl <int> 4, 4, 6, 6, 4, 6, 6, 4, 4, 6, 4, …
## $ displ <dbl> 2.5, 2.5, 4.2, 4.2, 2.5, 4.2, 3.8…
## $ fuel <chr> "Regular", "Regular", "Regular", …
## $ hwy <int> 17, 17, 13, 13, 17, 13, 21, 26, 2…
## $ cty <int> 18, 18, 13, 13, 16, 13, 14, 20, 2…Create summary tibble
annual.mpg <- vehicles %>% sample_frac(0.5) %>%
filter(cyl %in% 4:8) %>% group_by(year) %>%
summarize(hwy.avg = mean(hwy), hwy.sd = sd(hwy),
city.avg = mean(cty), city.sd = sd(cty)) %>%
arrange(desc(city.avg))Note: Here I used %in%, which works like inrange in Stata. You could alternately write two inequalities to achieve the same thing.
# Print annual.mpg
annual.mpg## # A tibble: 32 x 5
## year hwy.avg hwy.sd city.avg city.sd
## <int> <dbl> <dbl> <dbl> <dbl>
## 1 2015 28.6 5.42 20.6 4.78
## 2 2014 27.8 6.44 20.4 5.76
## 3 2013 27.3 6.02 20.0 5.78
## 4 2012 26.3 5.83 19.2 5.21
## 5 2011 25.7 5.36 18.8 4.86
## 6 2010 25.3 4.90 18.4 4.38
## 7 1985 22.8 6.21 17.7 4.70
## 8 2009 24.2 4.54 17.6 3.92
## 9 1986 22.4 5.82 17.5 4.48
## 10 1997 23.6 5.11 17.2 3.76
## # … with 22 more rowsSummarizing a data set with the summary() function
Although the tidyverse summarize() function is more powerful, often you just a want a quick look at summary statistics for the whole data set.
- You can easily do this with the base R summary() function, which produces summaries not just for data sets, but also for other R output like the results of a regression.
summary(wages)## wage schooling sex exper
## Min. : 1.69 Min. : 8 female:15 Min. : 3.000
## 1st Qu.: 44.07 1st Qu.:11 male :15 1st Qu.: 6.250
## Median : 160.96 Median :12 Median : 7.500
## Mean : 5941.66 Mean :12 Mean : 7.467
## 3rd Qu.: 1519.01 3rd Qu.:13 3rd Qu.: 8.000
## Max. :73659.06 Max. :15 Max. :13.000
Cleaning data
Common data cleaning tasks
There are a few data cleaning tasks that are pervasive in empirical work:
- Ensure columns have useful names
- Recoding variable values
- Addressing missing values
Renaming columns
Renaming columns is easily accommodated with the tidyverse rename() command.
Syntax:
mydataframe <- mydataframe %>% rename(NewVarName = OldVarName)To see rename() in action, let’s go back to the barro.lee.data educational data set we imported earlier:
Renaming columns example
Let’s look at columns 1 and 7 through 9:
glimpse(barro.lee.data[,c(1,7:9)], width = 50)## Observations: 1,898
## Variables: 4
## $ BLcode <dbl> 1, 1, 1, 1, 1, 1, 1, 1, 1, 1, 1,…
## $ lu <dbl> 80.68459, 81.05096, 82.61115, 80…
## $ lp <dbl> 17.563400, 17.018442, 14.313748,…
## $ lpc <dbl> 3.745905, 3.464397, 3.069391, 4.…See how these variable names are uninformative? Let’s use str() now.
str(barro.lee.data[,c(1,7:9)])## Classes 'tbl_df', 'tbl' and 'data.frame': 1898 obs. of 4 variables:
## $ BLcode: num 1 1 1 1 1 1 1 1 1 1 ...
## ..- attr(*, "label")= chr "Country Code"
## ..- attr(*, "format.stata")= chr "%8.0g"
## $ lu : num 80.7 81.1 82.6 80.9 73.6 ...
## ..- attr(*, "label")= chr "Percentage of No Schooling"
## ..- attr(*, "format.stata")= chr "%7.2f"
## $ lp : num 17.6 17 14.3 14.4 19.2 ...
## ..- attr(*, "label")= chr "Percentage of Primary"
## ..- attr(*, "format.stata")= chr "%7.2f"
## $ lpc : num 3.75 3.46 3.07 4.01 5.23 ...
## ..- attr(*, "label")= chr "Percentage of Primary Complete"
## ..- attr(*, "format.stata")= chr "%7.2f"Although the column names here are uninformative, the imported SAS file included variable labels saved as attributes, which can be seen using the more verbose str() function.
barro.lee.data <- barro.lee.data %>%
rename(countrycode = BLcode,
perc.noschool = lu,
perc.primary = lp,
perc.primary.complete = lpc)Now let’s look at the variable names again:
glimpse(barro.lee.data[,c(1,7:9)], width = 50)## Observations: 1,898
## Variables: 4
## $ countrycode <dbl> 1, 1, 1, 1, 1, 1,…
## $ perc.noschool <dbl> 80.68459, 81.0509…
## $ perc.primary <dbl> 17.563400, 17.018…
## $ perc.primary.complete <dbl> 3.745905, 3.46439…Recoding variables
Along with renaming variables, recoding variables is another integral part of data wrangling.
wages[1:4,"sex"] # Look at sex column## # A tibble: 4 x 1
## sex
## <fct>
## 1 female
## 2 female
## 3 male
## 4 malewages$sex <- wages$sex %>% recode("male"=0,
"female"=1) # recode
wages[1:4,"sex"] # Look at sex column## # A tibble: 4 x 1
## sex
## <dbl>
## 1 1
## 2 1
## 3 0
## 4 0Missing Values
Another problem characteristic of observational data is missing data. In R, the way to represent missing data is with the value NA.
- You can recode missing value that should be NA but are code using a different schema either by using brackets, or the tidyverse na_if() function.
## Replace 99-denoted missing data with NA
# bracket method
wages[wages$schooling==99,] <- NA
# tidyverse method
wages$schooling <- wages$schooling %>% na_if(99) You can check for (correctly-coded) missing-values using the is.na() function.
## Missing
wages[is.na(wages$wage),]## # A tibble: 3 x 4
## wage schooling sex exper
## <dbl> <int> <dbl> <int>
## 1 NA 14 1 8
## 2 NA 11 0 8
## 3 NA 10 0 8Note: R does not naturally support multiple types of missingness like other languages, although it’s possible to use the sjmisc package to do this.
Tidy data
Principles of tidy data
Rules for tidy data (from R for Data Science):
- Each variable must have its own column.
- Each observation must have its own row.
- Each value must have its own cell.
Tidy data tools in the tidyverse
There two main tidyverse verbs for making data tidy are:
gather(): reduces variable values are spread over multiples columns into a single column.
spread(): when multiple variables values are stored in the same columns, moves each variable into it’s own column.
Gathering data
If values for a single variable are spread across multiple columns (e.g. income for different years), gather moves this into single “values” column with a “key” column to identify what the different columns differentiated.
Syntax:
gather(data, key, value, columnstocombine)Gather example
earnings.panel## # A tibble: 7 x 3
## person y1999 y2000
## <chr> <dbl> <dbl>
## 1 Elsa 10 15
## 2 Mickey 20 28
## 3 Ariel 17 21
## 4 Gaston 19 19
## 5 Jasmine 32 35
## 6 Peter 22 29
## 7 Alice 11 15earnings.panel <- earnings.panel %>%
gather(key="year", value="wage",y1999:y2000)
earnings.panel## # A tibble: 14 x 3
## person year wage
## <chr> <chr> <dbl>
## 1 Elsa y1999 10
## 2 Mickey y1999 20
## 3 Ariel y1999 17
## 4 Gaston y1999 19
## 5 Jasmine y1999 32
## 6 Peter y1999 22
## 7 Alice y1999 11
## 8 Elsa y2000 15
## 9 Mickey y2000 28
## 10 Ariel y2000 21
## 11 Gaston y2000 19
## 12 Jasmine y2000 35
## 13 Peter y2000 29
## 14 Alice y2000 15Spreading data
Spread tackles the other major problem - that often times (particularly in longitudinal data) many variables are condensed into just a “key” (or indicator) column and a value column.
wages2## person indicator values
## 1 Elsa wage NA
## 2 Mickey wage 174.932480
## 3 Ariel wage 102.668810
## 4 Gaston wage 1.690623
## 5 Jasmine wage 2.166231
## 6 Peter wage 1192.371925
## 7 Alice wage 83.363705
## 8 Elsa wage NA
## 9 Mickey wage 174.932480
## 10 Ariel wage 102.668810
## 11 Gaston wage 1.690623
## 12 Jasmine wage 2.166231
## 13 Peter wage 1192.371925
## 14 Alice wage 83.363705
## 15 Elsa schooling 14.000000
## 16 Mickey schooling 12.000000
## 17 Ariel schooling 11.000000
## 18 Gaston schooling 11.000000
## 19 Jasmine schooling 14.000000
## 20 Peter schooling 12.000000
## 21 Alice schooling 11.000000
## 22 Elsa schooling 14.000000
## 23 Mickey schooling 12.000000
## 24 Ariel schooling 11.000000
## 25 Gaston schooling 11.000000
## 26 Jasmine schooling 14.000000
## 27 Peter schooling 12.000000
## 28 Alice schooling 11.000000
## 29 Elsa exper 8.000000
## 30 Mickey exper 5.000000
## 31 Ariel exper 7.000000
## 32 Gaston exper 8.000000
## 33 Jasmine exper 10.000000
## 34 Peter exper 8.000000
## 35 Alice exper 6.000000
## 36 Elsa exper 8.000000
## 37 Mickey exper 5.000000
## 38 Ariel exper 7.000000
## 39 Gaston exper 8.000000
## 40 Jasmine exper 10.000000
## 41 Peter exper 8.000000
## 42 Alice exper 6.000000wages2 %>% spread("indicator", "values")## person wage schooling exper
## 1 Elsa NA 14 8
## 2 Mickey 174.932480 12 5
## 3 Ariel 102.668810 11 7
## 4 Gaston 1.690623 11 8
## 5 Jasmine 2.166231 14 10
## 6 Peter 1192.371925 12 8
## 7 Alice 83.363705 11 6
## 8 Elsa NA 14 8
## 9 Mickey 174.932480 12 5
## 10 Ariel 102.668810 11 7
## 11 Gaston 1.690623 11 8
## 12 Jasmine 2.166231 14 10
## 13 Peter 1192.371925 12 8
## 14 Alice 83.363705 11 6